viewer
2011-07-19: P-values reported by the bin explorer dialogs are actual p-values, not log(p). Thanks to Marc-Chatenay-Lapointe, who reported the issue.
The Integrated Genomics Exploration Tools (IGET) viewer is a cross-platform (Qt) program for exploring the results from the motif, pathway, and interaction discovery tools available on this website and from Tavazoie Lab. In comparison to the PDF or HTML versions of output, information about the genes present in a given bin or intersection is more easily accessible.
The C++ parsing code for results is also available and may be used for result aggregation or further processing.
getting started
- download the program
- Download the compressed program package appropriate for your platform. Currently, binaries are available for Microsoft Windows (compatibility mode with Vista SP2 is recommended for W7) and Apple Mac platforms. The Qt source is also available for reference and use on other platforms.
- find results to view
- Obtain a set of results to be explored. If from the IGET website, extract the files from their archive. iPAGE results should be from the latest version with the new ipage_intersections file. Abnormal behavior may result from malformed files -- please use output files that have not been manipulated (and are from a trusted source).
- view results
- Explore your results (one tool at a time or all at once). Hover over bins and names for more information. Click on bins for gene information. Click on motif candidates for direct access to the regular expressions.
- offer feedback
- Are there features that would make this viewer more useful for you? Did you encounter any bugs? Let us know.
walkthrough
Click on the load button to choose an information file. It is assumed that the corresponding directory, information file _FIRE or _PAGE, contains the results.
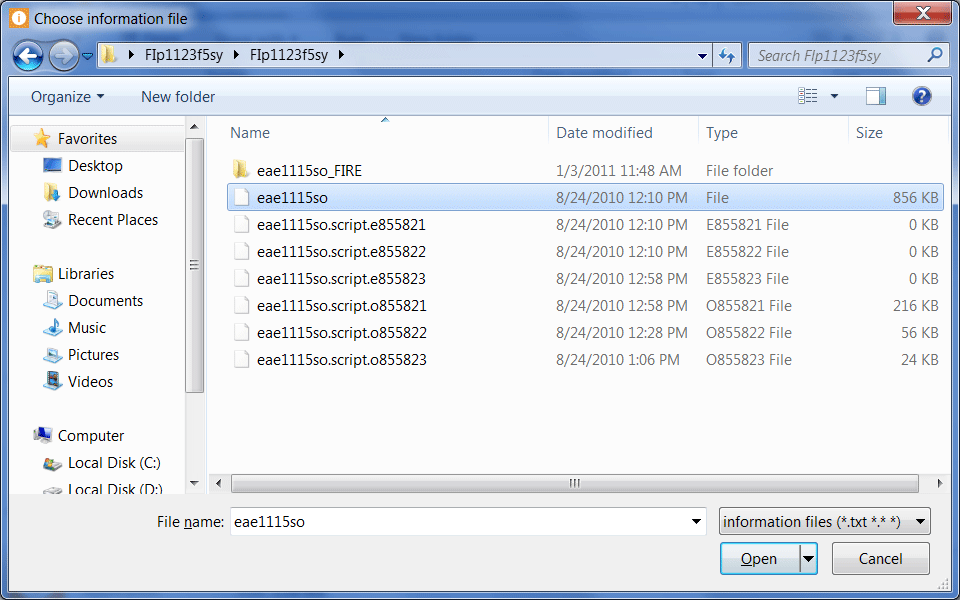
Locate and select an information file.
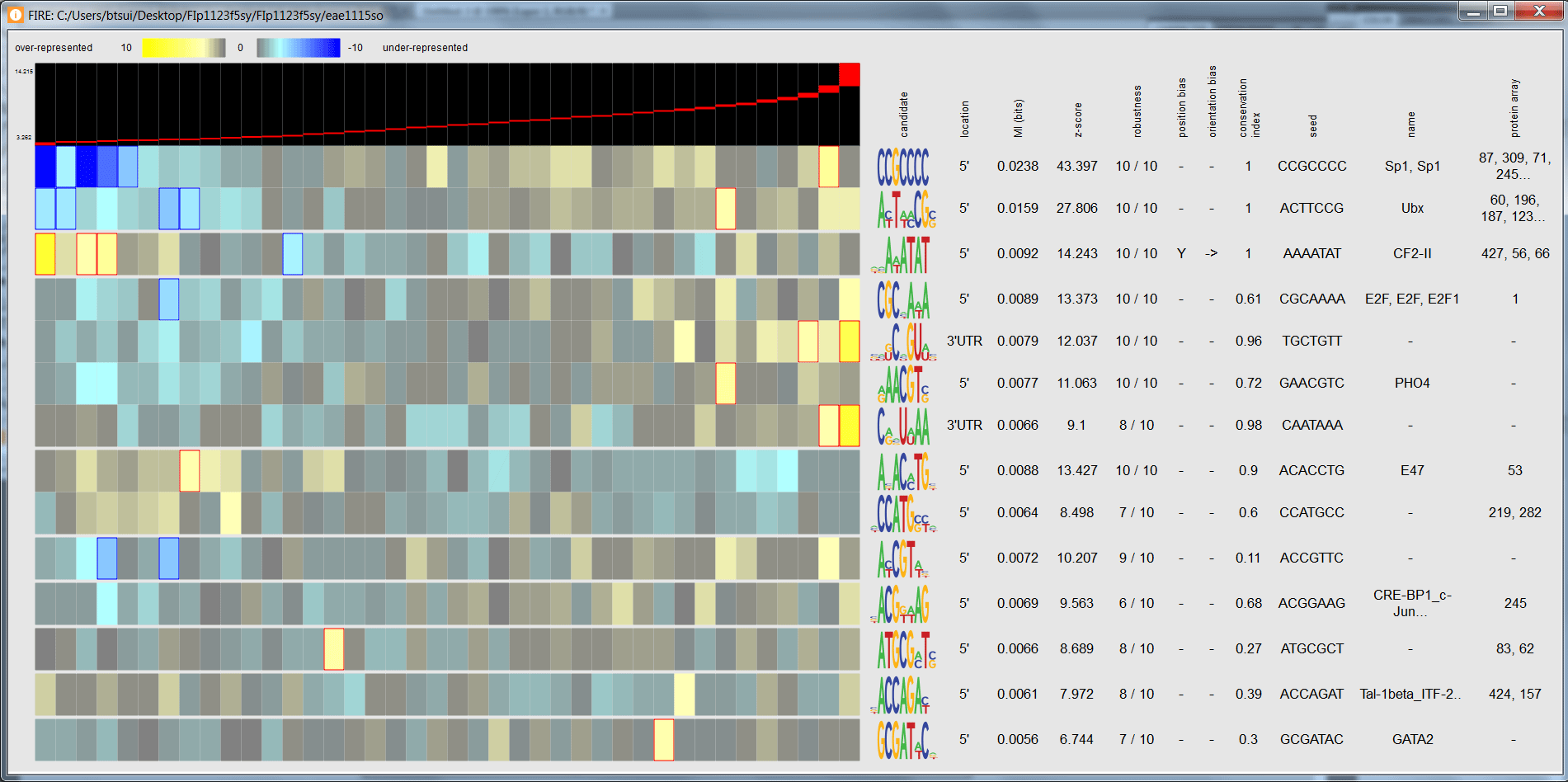
See an interactive display of the motif candidate summary.
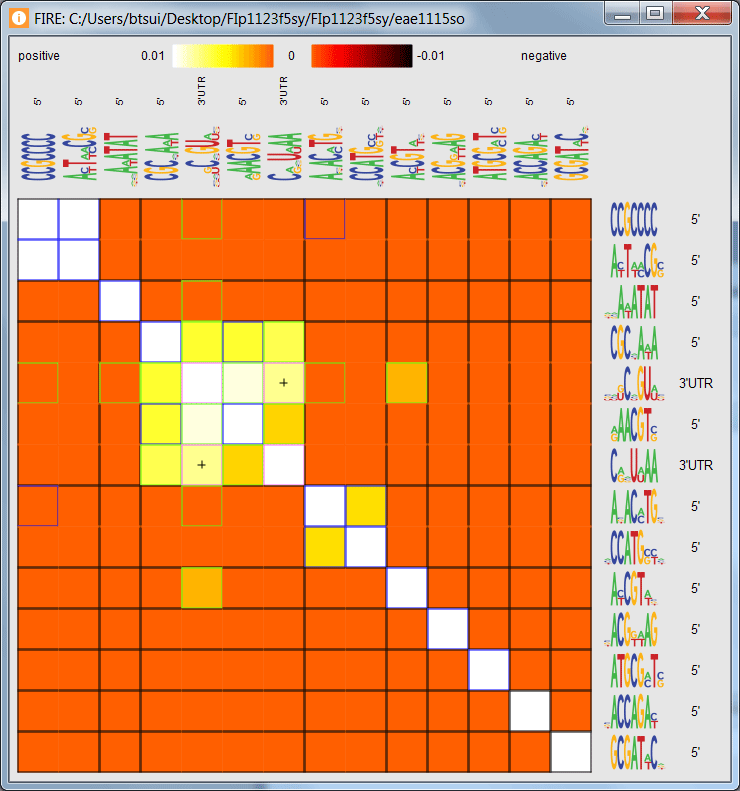
See an interactive display of the motif candidate interaction matrix.
Click on a bin at the top of the under/over-representation visualization to see the number and list of genes in a given expression bin.
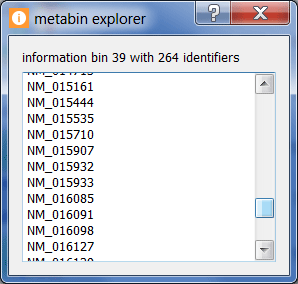
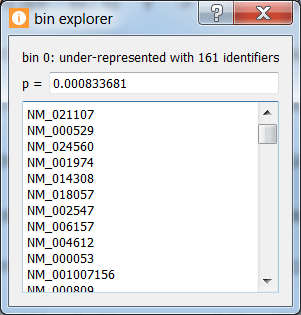
Click on a colored bin within the under/over-representation visualization for a given motif to see the number and list of genes in a given bin along with the bin's state and p-value.
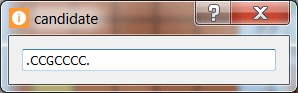
Click on motif candidate to see its regular expression.
Click on reset to choose another information file.
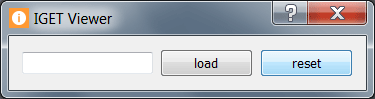
In the case of multiple results, select a subset or all the results for viewing.
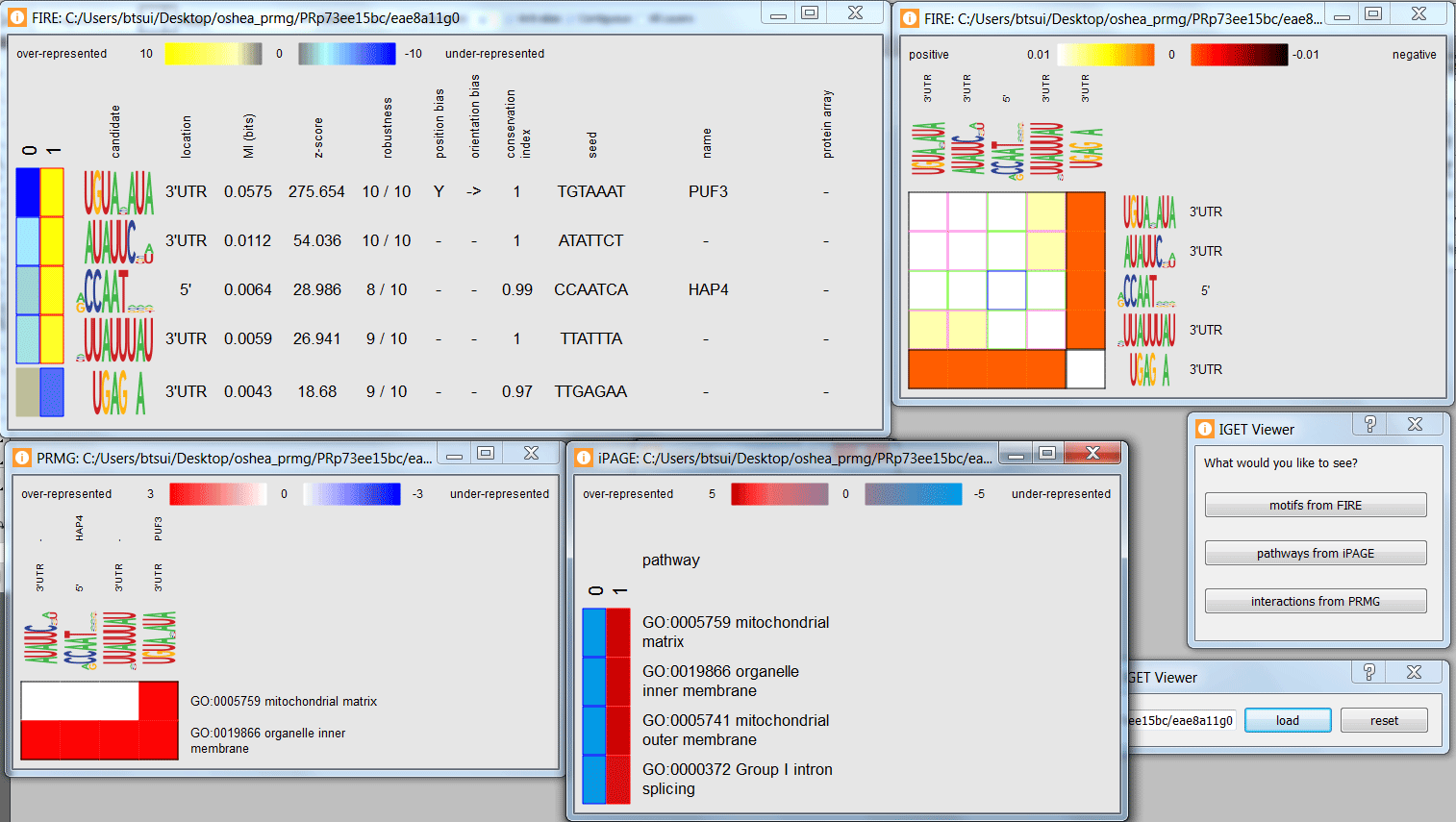
For example, with a high resolution display, one might examine the motifs, pathways, and interactions for a given information file at the same time.
parsing code
The C++ parsing code [h, cpp] is available for custom processing. The current version of the data structure description is dated August 20, 2010. It is recommended that one check the status variable in addition to motif and pathway counts to determine if parsing is successful (status = 1; motifs.count() > 0 or pathways.count() > 0).
contact us
Inquiries? Contact us by e-mail.
